Inspect: how it works ?
FAIR-Checker leverages semantic web technologies to check that metadata use standards
and recognized ontologies or controlled vocabularies.
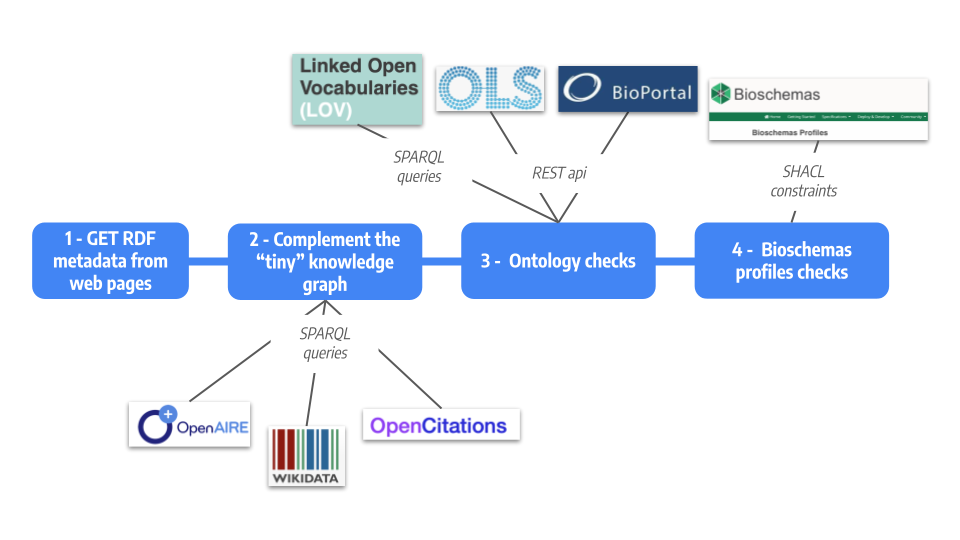
First, embedded semantic annotations are extracted from web pages, forming a minimal knowledge graph (Step 1). Then,
this minimal knowledge graph is completed based on already deployed knowledge graphs:
Datacite, OpenAire,
WikiData
(Step 2). Finally (Step 3), the resulting knowledge graph is tested to check that classes and properties are
recognized through Linked Open Vocabularies (LOV),
Ontology Lookup Service (OLS), or
Bioportal. The last
verification (Step 4) consists in validating the resource metadata against
Bioschemas community profiles.
The metadata assesment flow:
Resources
Gaignard, A., Rosnet, T., de Lamotte, F., Lefort, V., & Devignes, M. (2023).
FAIR-Checker: supporting digital resource findability and reuse with Knowledge Graphs and Semantic Web standards.
Journal of Biomedical Semantics, 14.
https://doi.org/10.1186/s13326-023-00289-5
Rosnet, Thomas, Lefort, Vincent, Devignes, Marie-Dominique, & Gaignard, Alban. (2021). FAIR-Checker, a web tool
to support the findability and reusability of digital life science resources. JOBIM (JOBIM), Paris.
https://doi.org/10.5281/zenodo.5914307
Gaignard, Alban, Rosnet, Thomas, de Lamotte, Frédéric, & Devignes, Marie-Dominique. (2021, June 4). Automatic
evaluation of FAIR metrics. Elixir All-Hands meeting.
https://doi.org/10.5281/zenodo.5914367
Funding
This project is funded by the French institute for Bioinformatics (IFB) through the PIA2 11-INBS-0013 grant.